 |
| Sargassum polycystum on a rocky shore at Tanjung Rimau, Sentosa (Author’s own collection). |
The macroalgae genus Sargassum C. Agardh (1820) comprises over 350 valid species globally [1], and are especially diverse and abundant in tropical and subtropical marine environments [2]. They are known to inhabit shallow reef flats and rocky bottoms as they often attach themselves to hard substrates, except for the two species, S. natans and S. fluitans which are holopelagic (floating Sargassum which reproduce on high seas) and form the Sargasso Sea in the North Atlantic Ocean.
The Sargasso Sea is vital for commercial fisheries and have an incredible potential in recreational fishing and ecotourism like whale watching where iconic species like the humpback whale are dependent on them. An estimated US$ 2.7 billion is generated annually from the ecosystem services they provide (in the Sargasso Sea alone), yet, the Sargassum habitat is still so poorly studied. There is however, hope for these areas to be protected and conserved as it has been recognised as an Essential Fish habitat.
What can you expect on local shores?
The where and when of Sargassum season:Mainly found on the Southern shores like Labrador on mainland Singapore, and on our offshore islands like Sentosa, St. John's Island, and Sister's Island and many more sites. The growth season begins in August and peaks in December to January, and eventually declines by March until only the perennial portions are left [8].
 |
| Left - Photo credit: Ria Tan from WildSingapore blog. Right: Imaged by Yip Zhi Ting. Thallus with stem, leaf and vesicle morphology of Singapore taxa: A) Sargassum aquifolium, B) S. swartzii, C) S. polycystum, D) S. cf. granuliferum, E) S. ilicifolium, F) S. sp. Scale bar: thallus = 5 cm; leaves = 1 cm; vesicles = 2 mm |
Table of Contents
Morphological features
Original description
The original description by Agardh (1824) in Systema algarum [7] documented Sargassum polycystum as having filiform stalks, lanceolate leaves with serrated margins and numerous vesicles (Fig. 1). The formal description of the species was however, not accompanied with any illustrations.
 |
| Fig. 1. Latin description of Sargassum polycystum (Agardh, C., 1824 in Systema algarum). |
Field identification
Studies in Singapore on the Sargassum community is still in infancy with only one study conducted by Low [8], which found only a total of five species despite 41 recorded in literature and ten deposited in the local herbaria. The Sargassum species present in Singapore can be differentiated and identified using the figure guide below (Fig. 2).
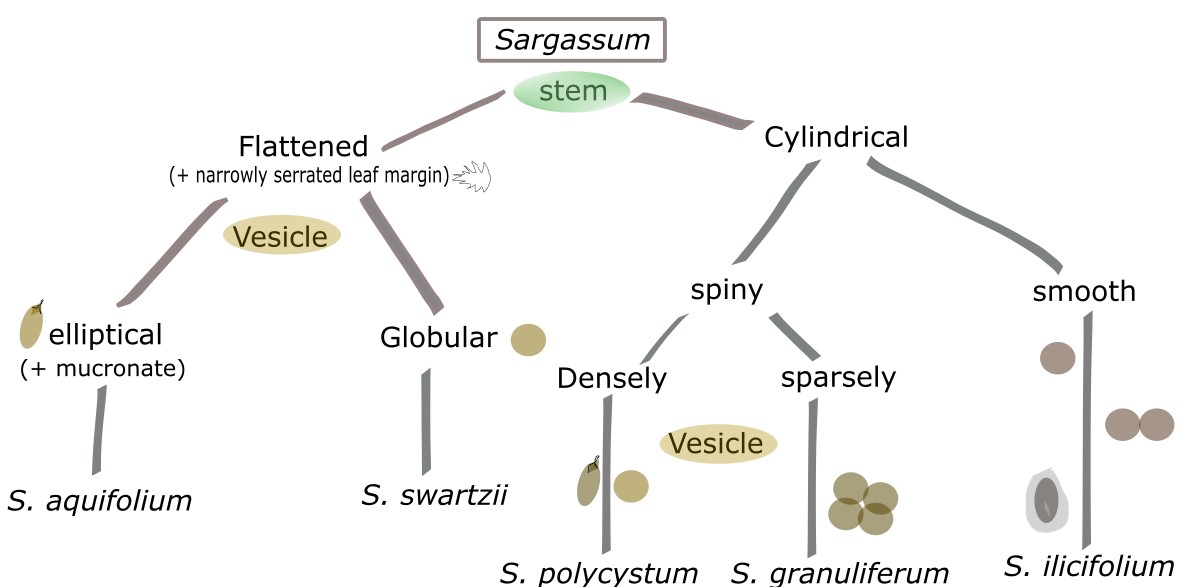 |
| Fig. 2. Morphological field guide for the identification of Singapore Sargassum species (Diagram by Yip Zhi Ting). |
Distinctive characteristics
Sargassum polycystum exhibits distinct morphology of variable thallus size depending on the exposure of water currents [5]. They are distinguished from other species by their discodial or conical holdfast, muricated stems with “y” shaped spine-link protuberances, leaves linear to lanceolate or oblong with irregularly and finely serrated margins, percurrent midrib, spherical or ovoid vesicles (smooth or mucronated) (Fig. 3) [5]. Male and female receptacles (reproductive parts) are located on different plants (unisexual) [5].
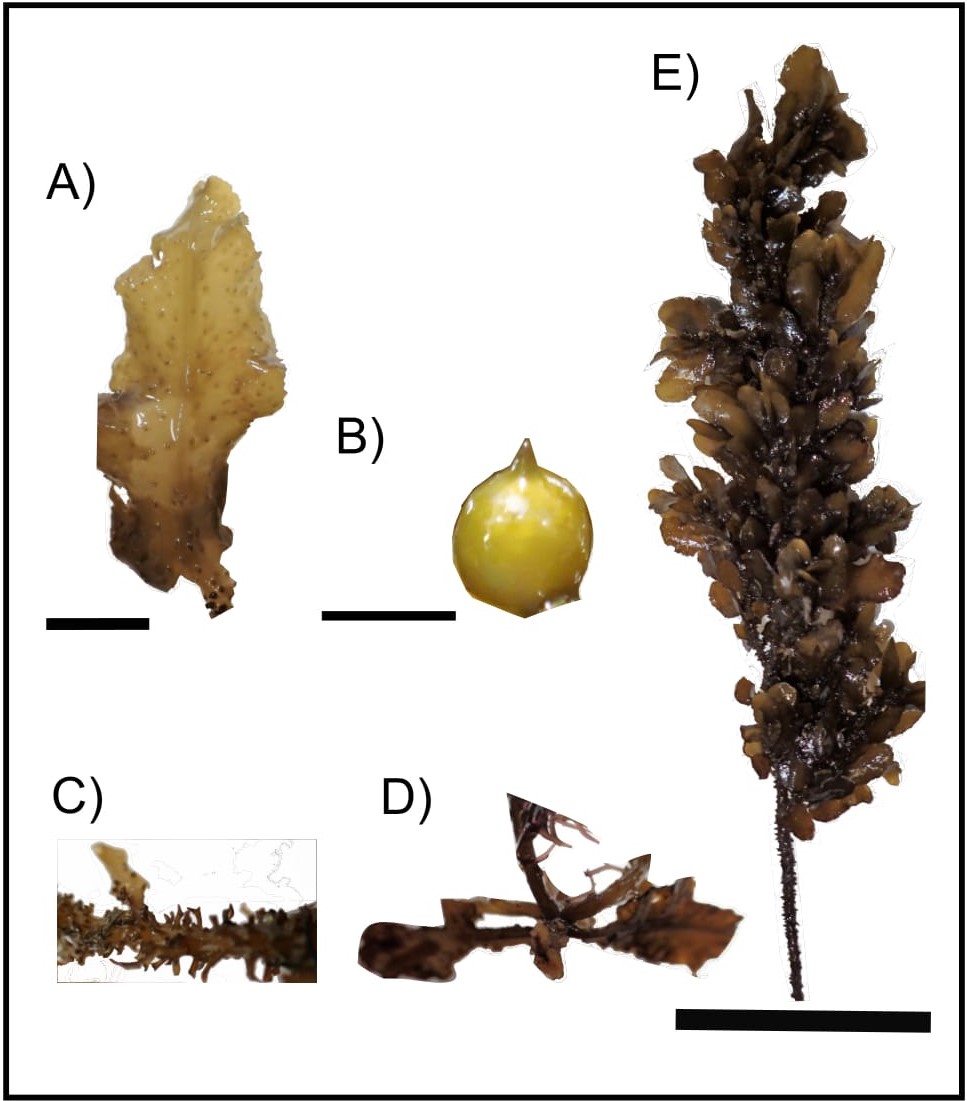 |
| Fig. 3. Morphology of the A) leaves, B) vesicles, C) stem, D) holdfast and E) thallus of Sargassum polycystum. Scale bar: thallus = 5 cm; leaves = 1 cm; vesicles = 2 mm. (Image by: Yip Zhi Ting) |
Biogeography
Global
Sargassum polycystum are mainly found in the Indo-pacific region and often on shallow reef flats and rocky bottom (Fig. 4) [5]. Sargassum polycystum of the Southeast Asian region specifically, has low genetic diversity [13]. Asexual propagation through vegetative methods in the form of drifting leaves with germlings, during a recent range expansion when they recolonised the newly flooded Sunda Shelf after the last Glacial Maximum promoted the occurrence of a homogeneous population [13].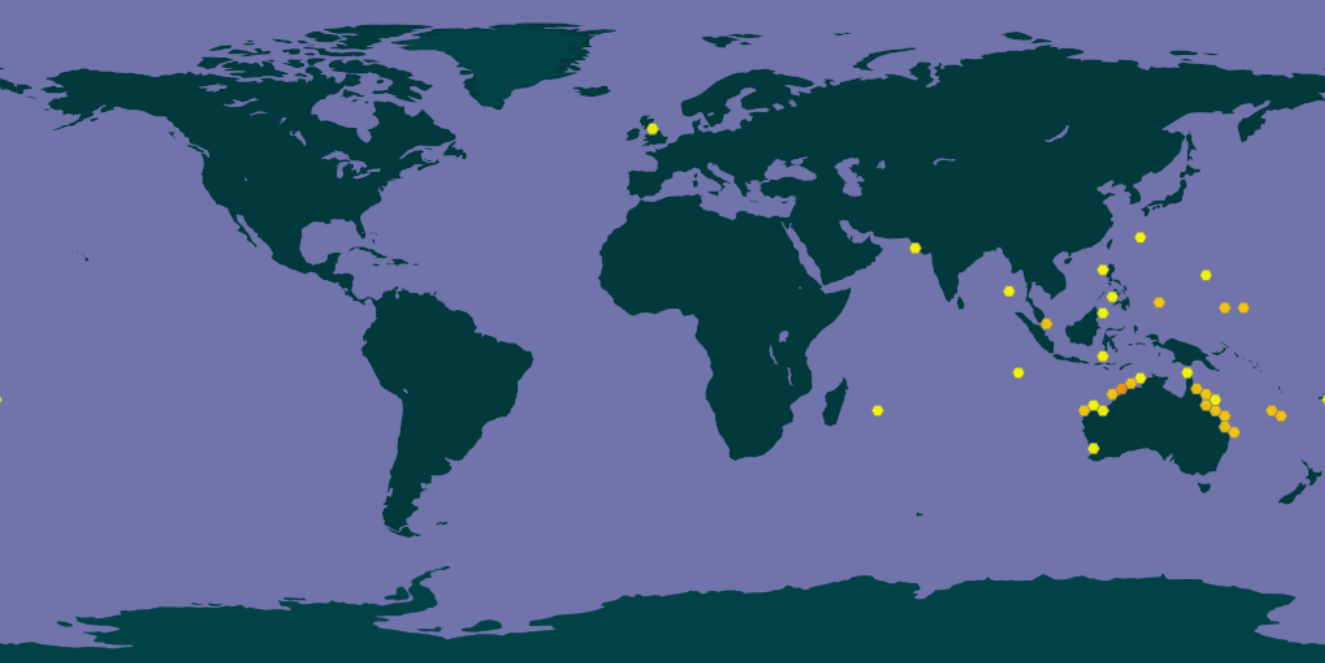 |
| Fig. 4. Global geographic distribution of Sargassum polycystum (GBIF Secretariat, 2017) [12]. Records are from published literature and herbarium input, hence it is by no means a complete representation of its full range. |
Local
Sargassum are mainly found on the southern shores and occasionally on the south-eastern shores (Fig. 5). They can be located on the reef flat (intertidal zone) or at the reef crest (subtidal).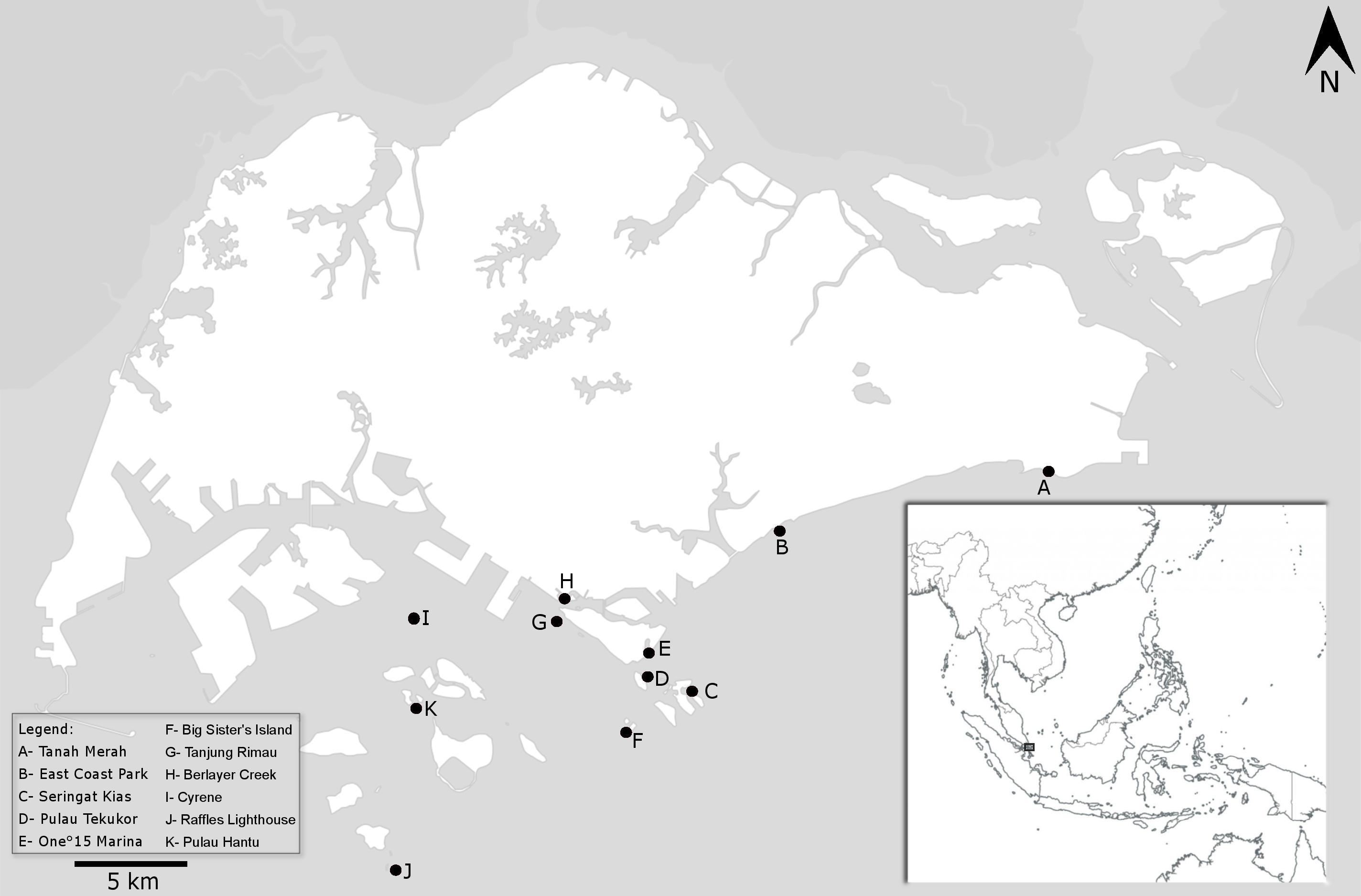 |
| Fig. 5. Map of Singapore with inset showing its location within Southeast Asia. Sightings were denoted by circles (Yip, 2017). |
Life on the shore
Biological and ecological interactions
Sargassum plays host to a myriad of marine organisms and in the Sargasso Sea where its boundaries are defined by major oceanic currents (gyres) instead of land mass, the Sargassum provides a food source, home, and shelter for many marine plants, shrimps, crabs, birds, fishes, turtles and whales [15, 16]. The Sargassum mats provide a safe refuge and nursery for juveniles until they are sufficiently large to venture into the open ocean [16]. Some have adapted and evolved to match the coloration and pattern of Sargassum for camouflage and there are now 10 endemic species living in the Sargasso Sea which includes a crab, nudibranch, Sargassum pipefish and the Sargassum fish (frog fish) [16]. Washed ashore Sargassum species and those found in the reef flat, serves as a food source for crabs, insects, and a rich diversity of tiny creatures (amphipods), which then feed shorebirds and other coastal animals in the intricate food web of interlinked communities [16].
 |
| Other marine fauna occasionally found on Sargassum in Singapore. Photos courtesy of Ria Tan from WildSingapore blog. |
PRECAUTION: Be wary of where you step in the inter-tidal zone, especially on mats of algae as venomous animals might lurk beneath.
Studies on species specific epifauna of the genus Sargassum in the region are still lacking. However, more is known of the epifauna communities on the Singapore species from a study by Low (2015) which sheds light on the delicate ecosystem they provide on our reef flats. According to Low, the most abundant epifauna observed were amphipods, copepods, gastropods, polychaetes, tanaids, isopods and bivalves, which constitutes more than 95% of total epifauna observed [8].
 |
| Common epifauna associated with Sargassum: A) Decapods, B) gastropods, C) isopods, D) amphipods, E) tanaids, F) polychaetes, G), anemone, H) polyclads. Photos A to H imaged by Gan Su Xuan except photo E (Source: Creative Commons by bathyporeia). |
Ecological interactions involving macroalgae are critical on coral reefs as coral-algae phase shifts follow environmental disturbances and may have considerable effects on reef coral community structure [23], diversity [24], and ecosystem services and function [25]. McCook et al. identified the following ways in which competition could arise between corals and macroalgae: overgrowth, shading, abrasion, allelopathic chemical effects and space pre-emption/ recruitment barrier [26].
 |
| Tall Sargassum growth taken at Lazarus Island. (Photo credit: Dr. Tay Y.C.) |
Life history and reproduction
The macroalgae genus Sargassum adopts a heteromorphic life history (distinct sexual haploid and asexual diploid stages) and oogamous fertilisation (union of mobile male and immobile female gametes) [14]. Sargassum polycystum is a dioecious macroalgae (male and female gametangia are formed on different individuals) [3] and sexual reproduction involves the fusion of motile sperm cells and sessile egg cells of the oogonium [17]. The motile young germlings developed from the fertilised zygotes are released from the female receptacles which then begin their journey for the search of a suitable substrate to adhere onto (Fig. 6) [17].
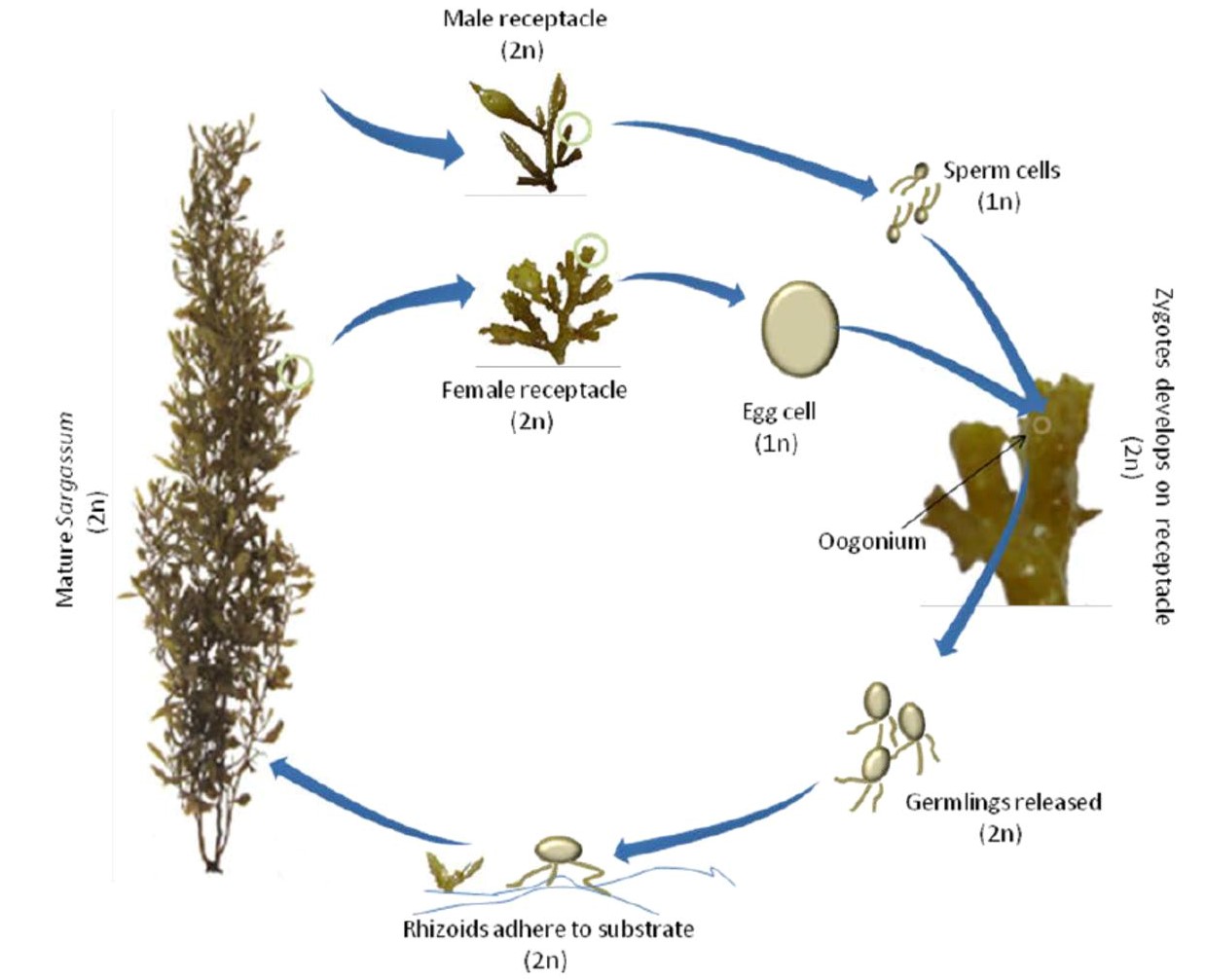 |
| Fig. 6. Life cycle of the macroalgae genus Sargassum (Nguyen, 2015). |
However, interestingly, Sargassum polycystum is the only species (excluding holopelagic species) from the genus so far that is known to also propagate asexually through vegetative methods (fragmentation) [13]. Fragments of the Sargassum polycystum (ramifying holdfast or stolons) were observed to have grown new main axes and subsequently reattaching to the substratum [13].
Economic importance
General usage
Sargassum polycystum serves a multitude of purpose from its usage as fertilizer, human food (click on link for tasty and nutritious recipe!), agricultural feed (fodder) and medicine, to controlling heavy metal (Pb, Cd) pollution and biogas production as dense stands makes them a good biomass source [11]. |
| Hijiki salad (Source: Creative Commons by Kirk K). |
Potential disease prevention and management
Sargassum polycystum possess potent antioxidant and anticancer properties from the isolated and extracted fucoidan, which is a natural sulfated polysaccharide and can be taken as a dietary supplement product [18]. Furthermore, extracts of Sargassum polycystum have been found to improve insulin sensitivity, blood sugar and lipid levels in type 2 diabetes rat model. The result has huge implications on the application as a potential insulin sensitizer for the management of type 2 diabetes [19]. In addition, it also has cardiovascular protective effects and anti-obesity properties which reduced body weight gain of the high cholesterol and high fat diet (HFC) rats [20].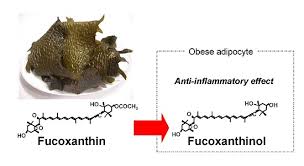 |
| Active compound Fucoxanthin and its effect on obese adipose tissue. (Source: Creative Commons Attribution License, Maeda, H., et al. Fucoxanthinol, Metabolite of Fucoxanthin, Improves Obesity-Induced Inflammation in Adipocyte Cells. Mar. Drugs 2015, 13, 4799-4813) |
Potential in the medical and pharmaceutical fields
Recent findings on the mediated synthesis of gold nanoparticles (AuNPs) using the seaweed Sargassum polycystum extract have raised interest in the biomedical field [21]. Biological synthesis reduces the dependency on toxic chemicals during the process and is critical for medical and biotechnological applications [21]. AuNPs are essential in the field due to their unique and tunable surface plasmon resonance (SPR) property which allows for a wide range of applications in biomedical science including drug delivery, tissue/tumour imaging, photo thermal therapy, immune chromatography, and identification of pathogens in clinical specimens [21]. Most of the available chemical processes for synthesis of gold nanoparticles (AuNPs) involve toxic chemicals that get adsorbed on the surface, leading to adverse effects in medical applications [21]. As such, there is a need to develop environmentally benign process for rapid synthesis of nanoparticles [21].The presence of alginate in Sargassum polycystum has also made them favourable in the biomedical industry. Alginate are sought after for their bio-compatibility and ease of gelation which is often manufactured into alginate hydrogels for purposes like wound healing, drug delivery and tissue engineering [22].
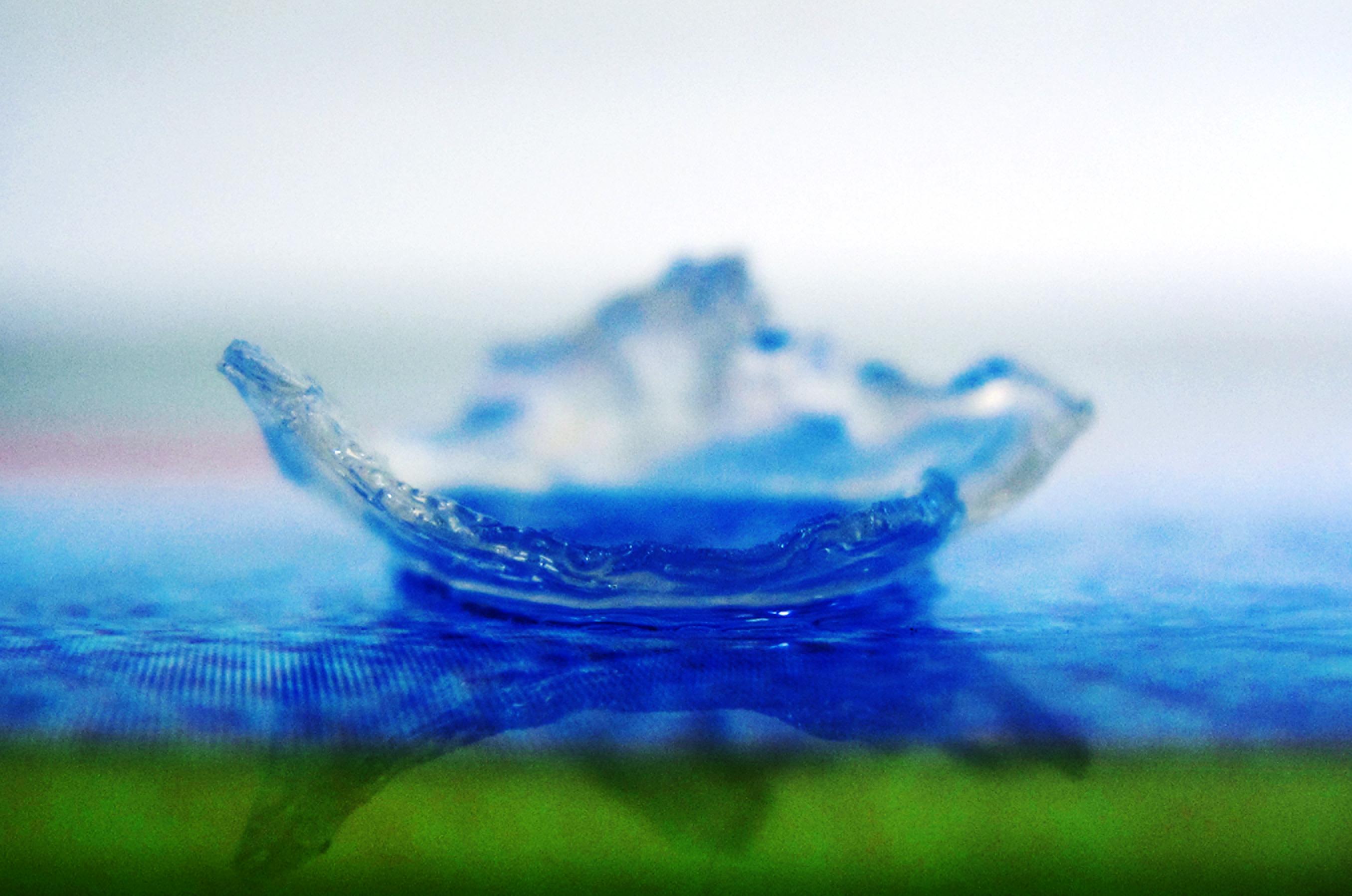 |
| A piece of transparent electrospun gelatin fibre-alginate hydrogel composite to be used as a scaffold to construct a cornea equivalent, which could be used for corneal transplantation when donor cornea is not available. This tissue-engineered cornea could restore vision to millions of people worldwide. (Source: Creative Commons by Engineering at Cambridge) |
Taxonomy and systematics
Origin of name
Etymology: Sargassum, a brown algae (seaweed) was a term coined by Portuguese sailors—which has even been attributed to Christopher Columbus (1492 expedition: first time someone reported crossing the Sargasso Sea). The origin of the specific epithet “polycystum” could not be traced back.
Scientific name: Sargassum polycystum
Vernacular name: コバモク (‘Kobamoku’) [9], Agar-agar koepan [10], Rough-stemmed sargassum (Australia) [11]
Known synonyms
Sargassum myriocystum f. horridula Grunow
Sargassum spinuligerum var. fissifolia f. humilis littoralis Grunow
Sargassum gaudichaudii Montagne 1842
Sargassum myriocystum J.Agardh 1848
Sargassum polycystum var. onustum J.Agardh 1848
Carpacanthus gaudichaudii (Montagne) Kützing 1849
Sargassum elegans Greville 1849
Sargassum brevifolium Greville 1849
Sargassum pygmaeum Kützing 1849
Sargassum ambiguum Sonder 1871
Sargassum polycystum var. horridulum Grunow 1874
Sargassum opacum J.Agardh 1889
Sargassum myriocystum var. elegans Grunow 1915
[12]
Type locality and specimens
Type locality: Sunda Strait, Indonesia. [7]
Holotype of Sargassum polycystum could not be located.
Syntypes: TCD 1108, 1109 (Dublin, Ireland) [5]
Higher taxonomic classification
Sargassum belongs to a higher order, Fucales which is part of the group of brown algae (Phaeophyta). They reside in the family Sargassaceae and Sargassum polycystum is nested within the revised Subgenus Sargassum and lower level Section Polycystae which consists of other members: S. herporhizum and S. plagiophyllum [5]. The ranking system, especially for below genus level have, in the past decade gone through many revisions due to the work in molecular phylogenetics [3], which revealed that classification based on morphology was usually not concordant with molecular phylogenetic analyses. Thus the rationale for revising ranks below genus level was to obtain a monophyly within the ranks (See Fig. 7).
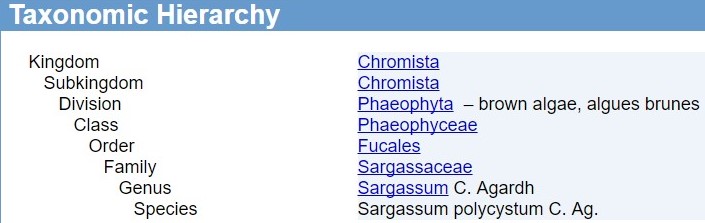 |
| Source: https://www.itis.gov/servlet/SingleRpt/SingleRpt?search_topic=TSN&search_value=203831#null |
Among the brown algae crown group (72 taxa selected in the study), the genus Sargassum (fully supported node of ML/BI=100/1.0) was most closely related to the genus Halidrys (Fig. 7A) [14]. The order Fucales which Sargassum belongs to is a sister group to the brown algae order Nemodermatales (Fig. 7A) [14]. The genus Sargassum was estimated to have originated approximately 30 MYA, and diversified about 10 MYA (Fig. 7B) along with the rapid diversification of the brown algae crown radiation (BACR) group [14]. The maximum likelihood (ML) phylogram in Fig. 7A was obtained from a concatenated data set of 10 genes with the sequence alignment conducted in MEGA version 4.0. An appropriate model (GTR + I + G) for the ML optimality criteria was chosen by applying partitioned model selection (AIC, BIC, LRT) and subsequently bootstrap for 1000 replicates. The data set was also analysed by Bayesian Inference using the program MrBayes V.3.1.2. The chronogram in Fig. 7B uses BEAST v.1.5.3 under a relaxed molecular clock method to infer a time-calibrated brown algae phylogeny. The partitioned data set was analysed with the GTR + I + G model, an uncorrelated (lognormal) clock model and Yule tree prior, and also calibrated using fossils from the Miocene deposits and Padina. Both trees were rooted with taxa Dictyotales, Sphacelariales and Syringodermatales as they have been shown to cluster as a sister group to the BACR [14].
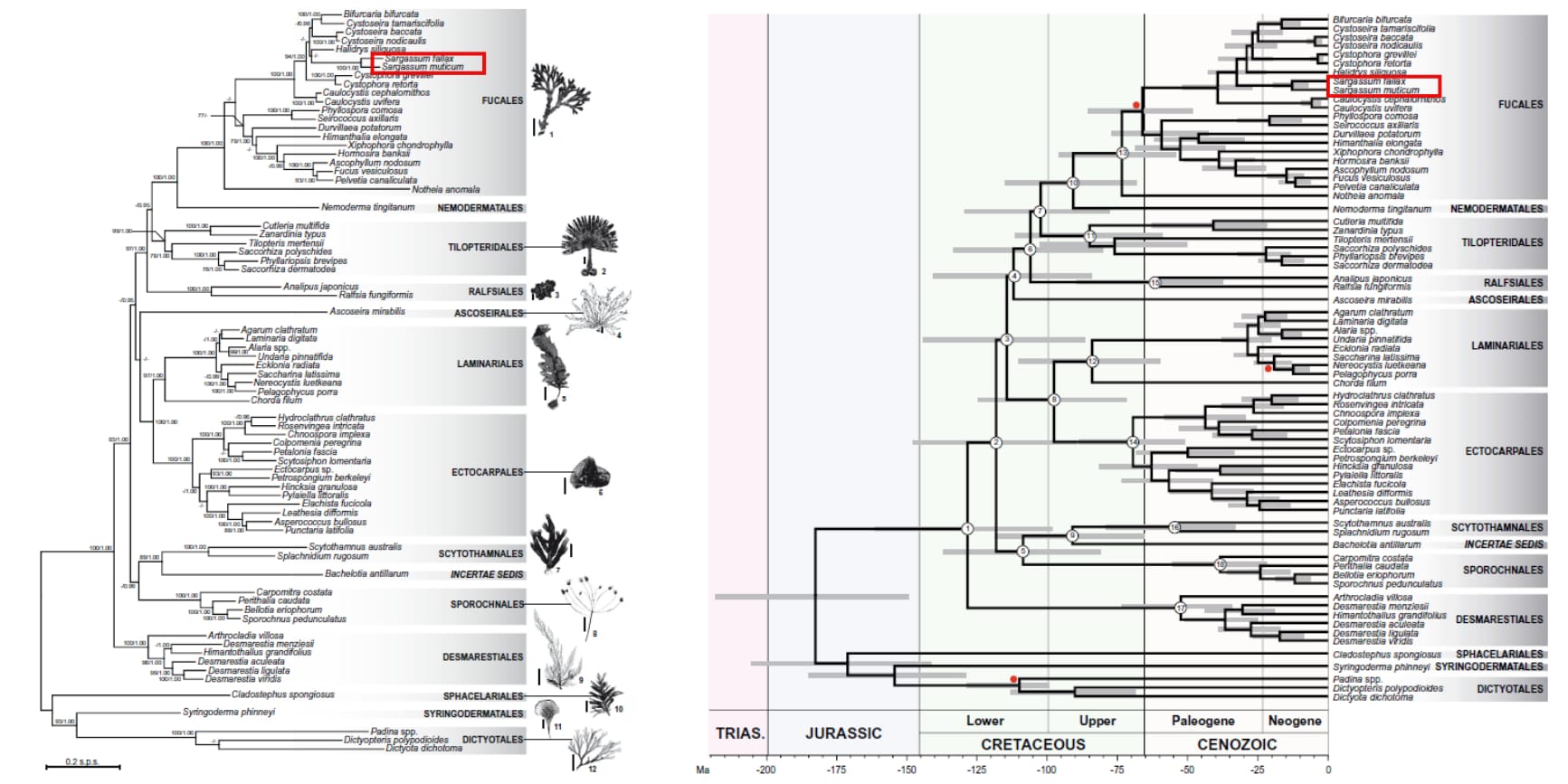 |
| Fig. 7. A) Maximum likelihood phylogram of the brown algae crown radiation (BACR) group consisting of 72 genes (10 genes partitioned data set). Branch label indicates the maximum likelihood bootstrap support and posterior probability in Bayesian inference. B) Chronogram of the BACR from a Bayesian relaxed molecular clock analysis using BEAST. The grey bars represent the 95% confidence interval (highest probability density) of the node age and the red circles are the clades that were time-calibrated using fossil records. Sargassum taxa were boxed in red. (Adapted from Silberfeld et al., 2010) |
Molecular phylogenetics
Diversity assessments on this genus of brown algae remains a challenge due to species identification using morphological features [3] as they exhibit high morphological plasticity in response to environmental changes, age and reproductive status [6]. Traditional classification of Sargassum which generated more than 1000 names with less than 40% currently accepted as valid taxa, were based on highly variable morphological characteristics [3]. The same species were misconstrued as a separate species due to a wrong interpretation of inter-specific variation [3]. However, the confusion created by the polymorphy of the genus was resolved by complementing traditional morphological characteristics with molecular analyses [4, 5] to better understand their relationship between species. The application of the Phylogenetic Species Concept sensu Mishler and Theriot in delimiting species of this genus would be most appropriate where the least inclusive monophyletic taxon is formally named. The usage of other species concept like the Biological and Hennigian species concept would be less relevant due to the exhibition of asexual reproduction by certain Sargassum species. Furthermore, the high morphological variability within each species might result in inaccurate species delimitation using the Phylogenetic Species Concept sensu Wheeler and Platnick, as it is heavily dependent on the initial population hypothesis.
Sargassum polycystum was originally placed in the section Malacocarpicae which was exclusively based on the presence of smooth and cylindrical receptacles arranged in cymes brush-like and pedicelate [3]. However, molecular phylogenetic analyses have shown that species classified in Malacocarpicae using these features were not genetically closely related in the phylogeny (Fig. 8) [3].
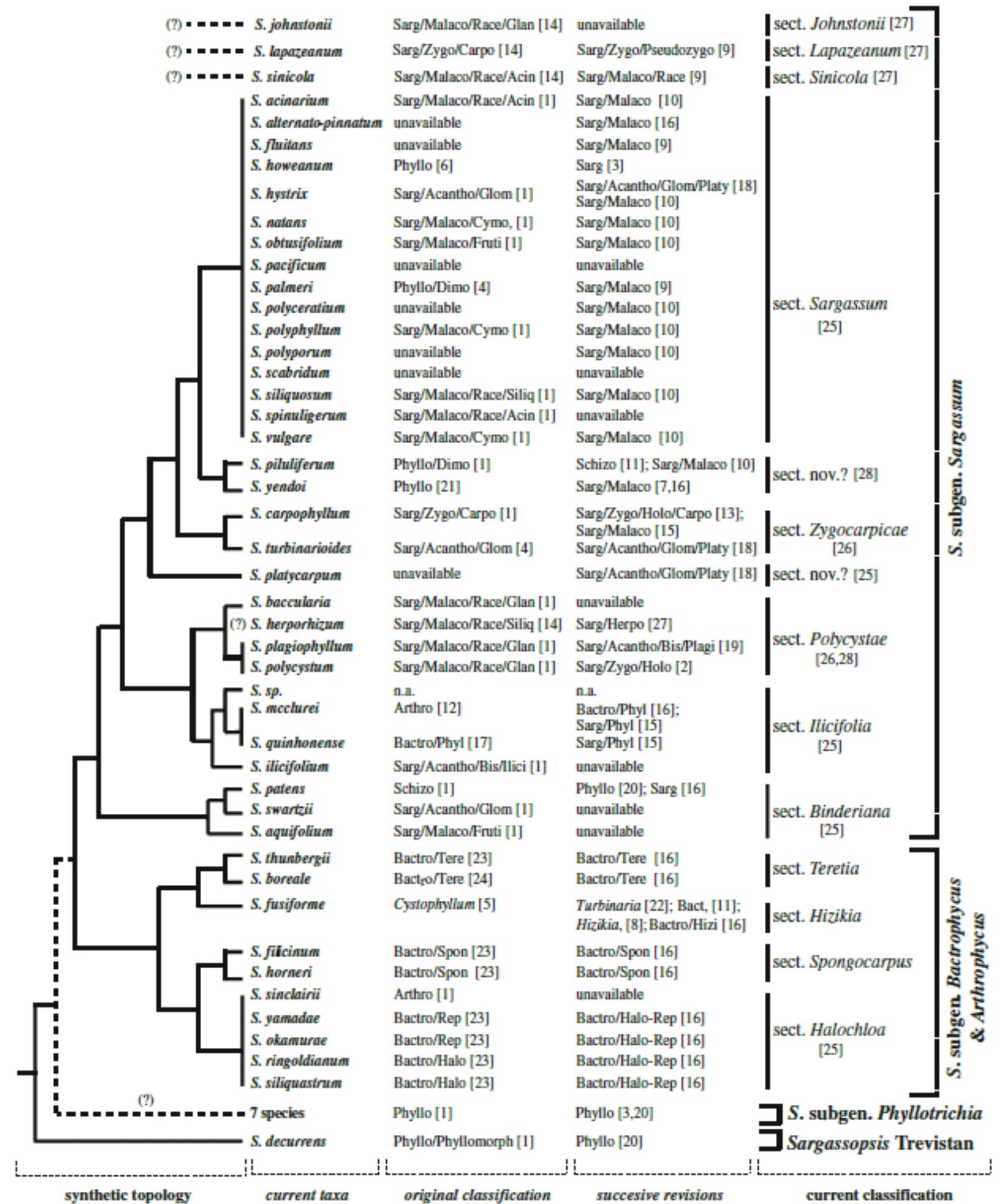 |
| Fig. 8. Taxonomy history synthesis for taxa sequenced since year 2000. Synthetic topology obtained from visual compilation of the ITS-2, rbc-LS and cox3 (Mattio & Payri, 2011). |
As such, recent revisions placed Sargassum polycystum in the section Polycystae distinguishable by their discoid or conical holdfast, stolon like horizontal branches which are smooth and cylindrical to flattened and branched, and densely clothed with leaves, vesicles and receptacles [5]. Molecular phylogenetics revealed that section Polycystae forms a monophyletic group, sister to the newly elevated section Ilicifoliae (Fig. 8) [3]. The topology of the sections within the subgenus Sargassum in the visually compiled tree (Fig. 8) were congruent with the concatenated (ITS-2, rbc-LS, cox3) neighbour-joining tree [4] and nodes for sections were well-supported by high bootstrap values [NJ/ MP/ ML: 100/99/100] (Fig. 9A). Furthermore, the topology of the tree obtained by a different optimality criteria, maximum likelihood (Fig. 9B) [29] was congruent with the neighbour-joining tree and also strongly supported with high bootstrap values [MP/ ML/ BI: 99/100/1], despite a difference in alignment method (Sequences aligned with BioEdit sequence alignment editor in Fig. 9A; 'auto' default setting in MAFFT 7.304 in Fig. 9B). Both trees were rooted with the outgroup taxa Turbinaria which was recovered as a sister clade to the genus Sargassum in the phylogeny.
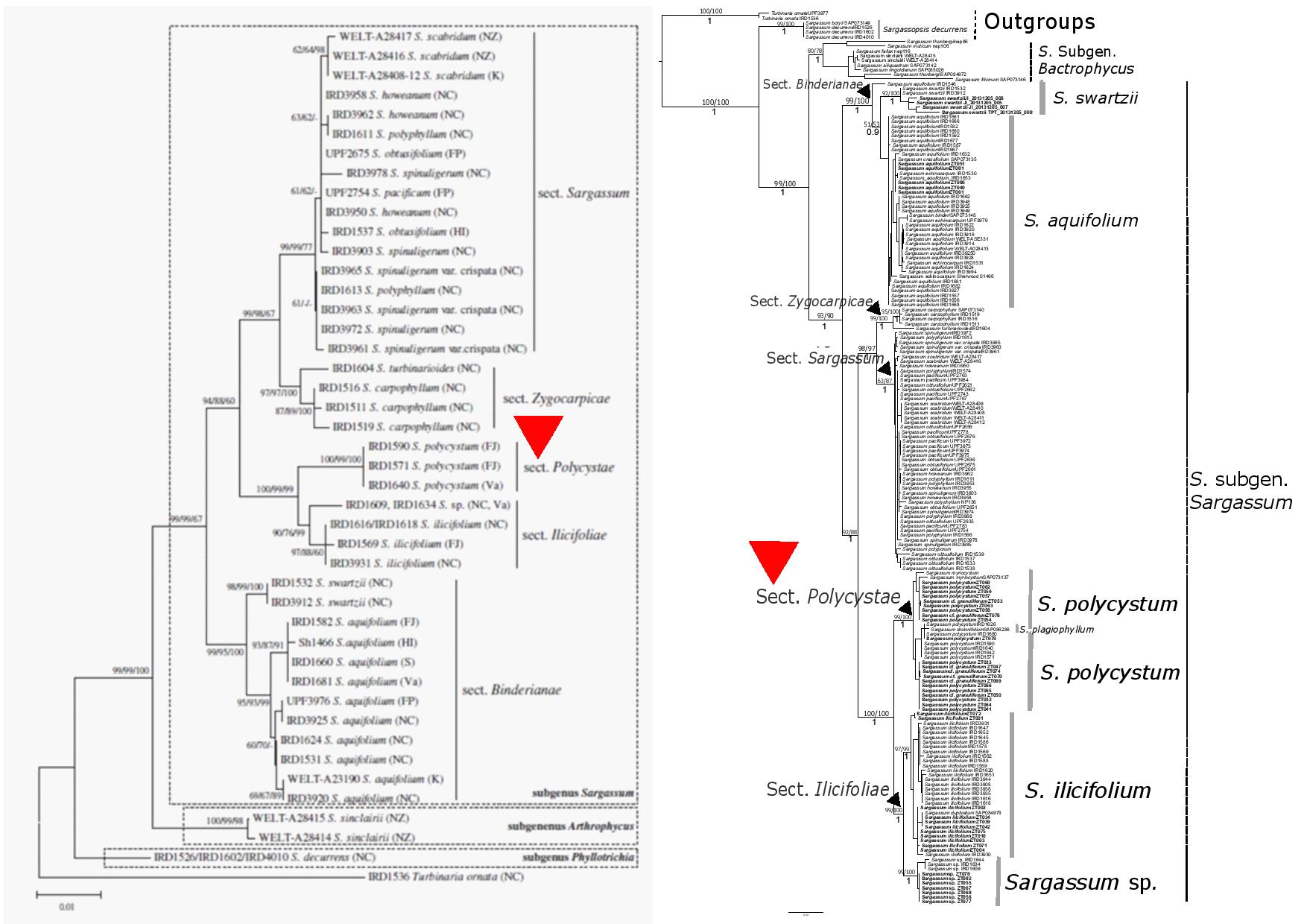 |
| Fig. 9. A) Neighbour- joining tree represented for concatenated dataset of ITS-2, cox3 and rbc-LS alignment. Bootstrap proportions for neighbour-joining/ maximum parsimony/ maximum likelihood indicated at each node (Mattio & Payri, 2009). B) Phylogenetic reconstruction of Sargassum (Phaeophyceae) with Turbinaria as outgroup, based on a maximum likelihood (ML) analysis of the concatenated ITS-2 + rbcLS + cox3 alignment. Bootstrap proportions indicated for maximum parsimony (MP)/ maximum likelihood (ML) when >0.5 and posterior probabilities for Bayesian inference (BI) when >0.8 for nodes at the species level and above (Yip, 2017). |
On our shores, Sargassum polycystum can sometimes be mistaken as another species, Sargassum granuliferum due to similarities in stem morphology (cylindrical and spiny). However, they are distinct from each other based on vesicles abundance and leaf morphology (refer to Fig. 2; Fig. 10). Molecular analyses using the three genes, ITS-2, cox3 and rbcLS have shown that the two species do not form separate monophyletic groups within this section. Sargassum granuliferum sequences are instead nested within S. polycystum sequences. This could be attributed to the lack of signal from molecular markers which can be overcome with more genetic markers to support their delimitation and facilitate reliable species identification [29].
 |
| Fig. 10. Section Polycystae of the maximum likelihood tree with Sargassum granuliferum sequences boxed in red, accompanied with morphological features of S. granuliferum below the tree. |
Conservation
The status of Sargassum polycystum has not been evaluated in both IUCN Red List and CITES [27, 28]. However, climate change has been predicted to affect the spatial distribution range of certain species of Sargassum like S. horneri, where its potential distribution was predicted to shift to higher latitudes by year 2100 in response to warming waters (Fig. 11) [30]. Temperate species at the southern limit of their geographical range have also been predicted to be replaced by subtropical and tropical species [30]. Although no studies have been carried out on S. polycystum to assess the effect of global warming on its population and shifts in geographical range, it is probable that its range might shift and affect ecosystems.
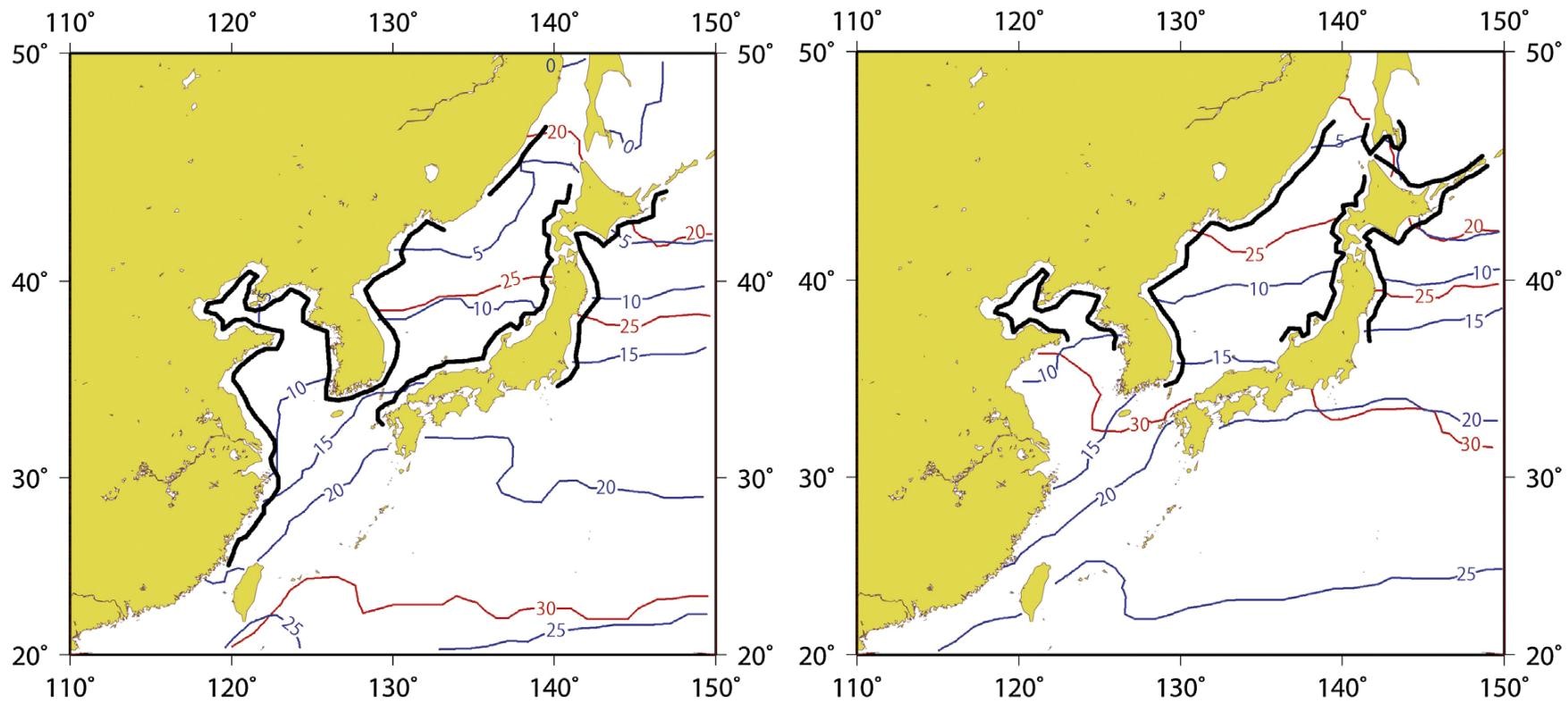 |
| Fig. 11. Potential distributions of S. horneri (black solid lines) estimated from distribution ranges of water temperature in February (minimum water temperature in a year) and in August (maximum temperature in a year), 2050 (left) and 2100 (right) based on simulation results of 12 models under A2 scenario. Source: Komatsu et al. (2014) under Creative Commons License. |
References
[1] M.D. Guiry in Guiry, M.D. & Guiry, G.M. 2017. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. http://www.algaebase.org; searched on 12 October 2017.
[2] Phillips, N. (1995). Biogeography of Sargassum (Phaeophyta) in the Pacific basin. In: Abbott, I.A. (Ed.), Taxonomy of Economic Seaweeds with Reference to Some Pacific Species Vol. V. California Sea Grant College, La Jolla, pp. 107–144.
[3] Mattio, L., Payri, C.E. (2011).190 years of Sargassum taxonomy, facing the advent of DNA phylogenies. Bot Rev 77(1):31-70.
[4] Mattio L., Payri C.E. (2009). Taxonomic revision of Sargassum species (Fucales, Phaeophyceae) from New Caledonia based on morphological and molecular analyses. J Phycol 45:1374- 1388.
[5] Mattio, L., Payri C.E., Verlaque, M. (2009). Taxonomic revision and geographic distribution of the subgenus Sargassum (Fucales, Phaeophyceae) in the Western and Central Pacific islands based on
morphological and molecular analyses. J. Phycol. 45: 1213-1227.
[6] Kilar JA, Hanisak MD, Yoshida T (1992) On the expression of phenotypic variability: why is Sargassum so taxonomically difficult? In Abbott, I. A. [Ed.] Taxonomy of Economic Seaweeds. Vol. 3. California Sea Grant College Program, La Jolla, California, pp 95–117.
[7] Agardh, C.A. (1824). Systema algarum. pp. [i]-xxxvii, [1]-312. Lundae [Lund]: Literis Berlingianis [Berling].
[8] Low, J. K. Y. (2015). Sargassum on Singapore's reefs. PhD. National University of Singapore, 1–183 pp.
[9] Japan Agency for Marine-Earth Science and Technology. (2009 onwards). Biological Information System for Marine Life (BISMaL). Accessed on 2017-03-02.
[10] Kirby, R.H. (1953). Seaweeds in Commerce. pp. 157pp. London: Her Majesty's Stationery Office.
[11] Trono, G.C. Jr. (2001). Seaweeds. p. 19-99. In Carpenter, K.E. and V.H. Niem (eds.), The Living Marine Resources of the Western Central Pacific, Vol. 1. FAO Species Identification Guide for Fishery Purposes. FAO, Rome. 686 p.
[12] Sargassum polycystum C.Agardh, 1824 in GBIF Secretariat (2017). GBIF Backbone Taxonomy. Checklist Dataset https://doi.org/10.5072/hufs9m accessed via GBIF.org on 2017-10-12.
[13] Chan, S.W., Cheang, C.C., Chirapart, A., Gerung, G., Tharith, C. et al. (2013). Homogeneous Population of the Brown Alga Sargassum polycystum in Southeast Asia: Possible Role of Recent Expansion and Asexual Propagation. PLoS ONE 8(10): e77662.
[14] Silberfeld, T., Leigh, J.W., Verbruggen, H., Cruaud, C., de Reviers, B., Rousseau, F. (2010). A multi-locus time-calibrated phylogeny of the brown algae (Heterokonta, Ochrophyta, Phaeophyceae): Investigating the evolutionary nature of the "brown algal crown radiation". Molecular Phylogenetics and Evolution, 56(2): 659- 74.
[15] Casazza, T.L., Ross, S.W. (2008). Fishes associated with pelagic Sargassum and open water lacking Sargassum in the Gulf Stream off North Carolina. Fish Bull, 106: 348– 363.
[16] Trott, T.M., McKenna, S.A., Pitt, J.M., Hemphill, A., Ming, F.W., Rouja, P., Gjerde, K.M., Causey, B., Earle, S.A. (2010). Efforts to enhance protection of the Sargasso Sea. Proceedings of the 63rd Gulf and Caribbean Fisheries Institute. Nov 1–5, 2010, San Juan, Puerto Rico, pp 282–286.1
[17] Nguyen, V. T. (2015). Seaweed diversity in Vietnam, with an emphasis on the brown algal genus Sargassum. PhD. Ghent University, 1-199 pp.
[18] Palanisamy, S., Vinosha, M., Marudhupandi, T., Rajasekar, P., Prabhu, N.M. (2017). Isolation of fucoidan from Sargassum polycystum brown algae: Structural characterization, in vitro antioxidant and anticancer activity. International Journal of Biological Macromolecules, vol. 102: 405-412.
[19] Motshakeri, M., Ebrahimi, M., Goh, Y.M., Matanjun, P., Mohamed, S. (2013). Sargassum polycystum reduces hyperglycaemia, dyslipidaemia and oxidative stress via increasing insulin sensitivity in a rat model of type 2 diabetes. Journal of the Science of Food and Agriculture, 93(7):1772-8.
[20] Matanjun, P., Mohamed, S., Muhammad, K., Mustapha, N. M. (2010). Comparison of cardiovascular protective effects of tropical seaweeds, Kappaphycus alvarezii, Caulerpa lentillifera, and Sargassum polycystum, on high-cholesterol/high-fat diet in rats. Journal of Medicinal Food, 13(4):792-800.
[21] Sivaraj, R., Priya, S.V.R., Rajiv, P., Rajendran, V. (2015). Sargassum Polycystum C.Agardh Mediated Synthesis of Gold Nanoparticles Assessing its Characteristics and its Activity against Water Borne
Pathogens. J Nanomed Nanotechnol 6:280. doi:10.4172/2157-7439.1000280
[22] Lee, K.Y., Mooney, D.J. (2012). Alginate: properties and biomedical applications. Progress in Polymer Science, 37(1): 106–126.
[23] Hughes, T.P. (1994). Catastrophes, phase shifts, and large-scale degradation of a Caribbean coral reef. Science 265, 1547-1551.
[24] Jones, G.P., McCormick, M.I., Srinivasan, M., Eagle, J.V. (2004). Coral decline threatens fish biodiversity in marine reserves. Proceedings of the National Academy of Sciences of the United States of America, 101(21), 8251-8253.
[25] Bellwood, D.R., Hughes, T.P., Folke, C., Nyström, M. (2004). Confronting the coral reef crisis. Nature 429, 827-833.
[26] McCook, L.J., Jompa, J., Diaz-Pulido, G. (2001). Competition between corals and algae on coral reefs: a review of evidence and mechanisms. Coral Reefs, 19: 400–417.
[27] CITES, UNEP-WCMC. (2016). The Checklist of CITES Species Website. Appendices I, II and III valid from 10 March 2016. CITES Secretariat, Geneva, Switzerland. Compiled by UNEP-WCMC, Cambridge, UK. https://www.cites.org/eng/app/appendices.php [Accessed 23/06/2016].
[28] IUCN. (2017). The IUCN Red List of Threatened Species. Version 2017-1. Downloaded on 18 May 2017.
[29] Yip, Z.T. (2017). Diversity and phylogeny of Sargassum in Singapore. UROPs. National University of Singapore.
[30] Komatsu, T., Fukuda, M., Mikami, A., Mizuno, S., Kantachumpoo, A., Tanoue, H., Kawamiya, M. (2014). Possible change in distribution of seaweed, Sargassum horneri, in northeast Asia under A2 scenario of global warming and consequent effect on some fish. Marine Pollution Bulletin, 85(2):317-24.